About RFE45Cancer Project
Project Overview
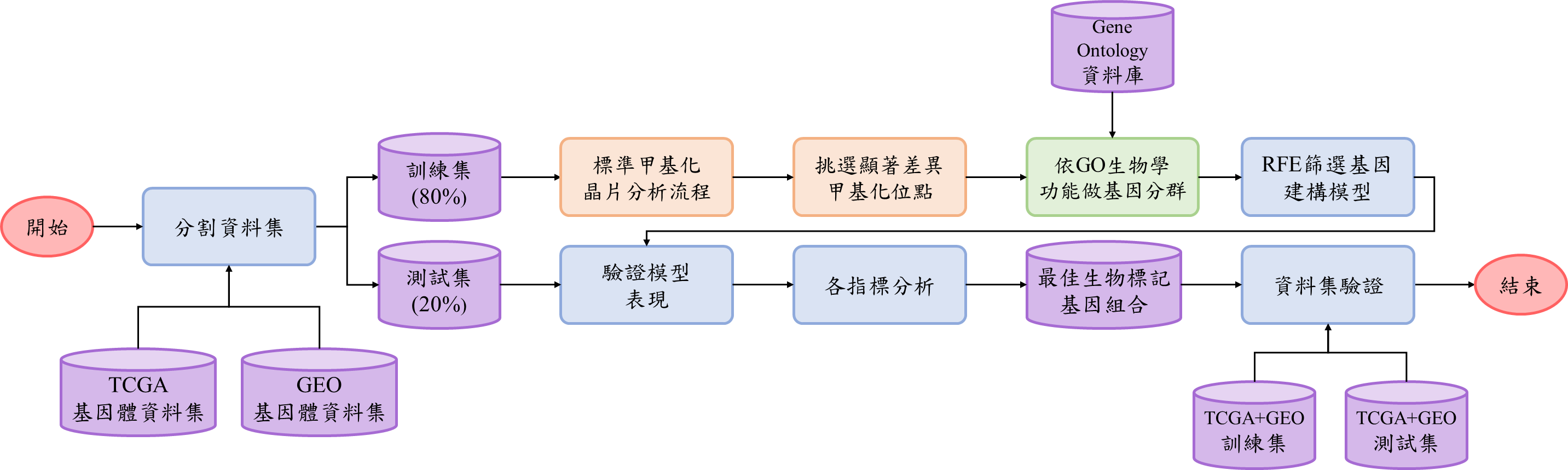
RFE45Cancer is a comprehensive research initiative focused on analyzing DNA methylation patterns across multiple cancer types to identify epigenetic biomarkers with diagnostic and prognostic potential. Our project integrates data from both The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) to provide robust, cross-validated findings.
Using advanced bioinformatics approaches and machine learning algorithms, particularly Recursive Feature Elimination (RFE), we identify gene signatures that can differentiate between cancer and normal tissues with high accuracy.
Our Methodology
Data Integration
Combining multiple datasets from TCGA and GEO databases to ensure robust findings across diverse patient populations.Methylation Analysis
Comprehensive preprocessing of methylation data using ChAMP package, including quality control, normalization, and batch effect correction.
Feature Selection
Application of Recursive Feature Elimination (RFE) to identify the most predictive DNA methylation markers.
Machine Learning
Ensemble-based voting classifiers combining Random Forest, SVM, and Gradient Boosting to improve classification stability and accuracy.
Validation
Rigorous cross-validation techniques to ensure the generalizability of our findings.
Project Process

Project Significance
DNA methylation alterations are crucial epigenetic changes in cancer development and progression. Our research aims to identify methylation signatures that can:
- Serve as early diagnostic biomarkers for cancer detection
- Predict patient outcomes and treatment responses
- Provide insights into cancer biology and potential therapeutic targets
- Contribute to the development of non-invasive liquid biopsy approaches
By focusing on consistent methylation patterns across multiple datasets, we enhance the reliability and clinical applicability of our findings.
